CMap COMPARATIVE MAP VIEWER
The CMap Comparative Map Viewer allows a user to view and compare maps between and among species. See HELP for more information.
 |
Save link
* Bookmarks for this page will fail after this session expires. Use the "Save Link" button to create a permanent link.
General options
** Eliminate Orphans - Remove comparison maps that don't have correspondences to a reference map.
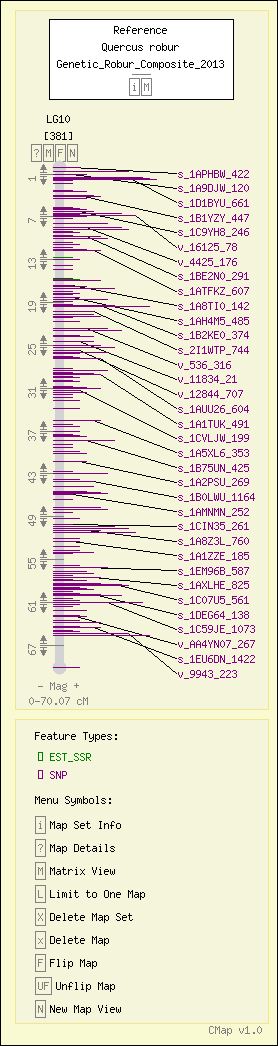
Map Details
| Map Type: | Genetic [ View Map Type Info ] | ||||||
|---|---|---|---|---|---|---|---|
| Map Set Name: | Quercus robur - Genetic_Robur_Composite_2013 [ View Map Set Info ] | ||||||
| Map Name: | LG10 | ||||||
| Map Start: | 0.00 cM | ||||||
| Map Stop: | 70.07 cM | ||||||
| Features by Type: |
|
Map Features
| [ Download Map Data ]
[ Download Feature Correspondence Data ] |
||||||||
| Quercus robur - Genetic_Robur_Composite_2013 - LG10 (Click headers to resort) |
Comparative Maps | |||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Type | Position (cM) | Map | Feature | Type | Position | Evidence | Actions |
| s_1AKX9X_1015 | SNP | 6.66 | No other positions | |||||
| s_1A6P85_230 | SNP | 6.55 | No other positions | |||||
| s_1A1M3H_1148 | SNP | 6.55 | No other positions | |||||
| s_1B1V8C_1374 | SNP | 20.23 | No other positions | |||||
| s_1A5XL6_353 | SNP | 35.50 | No other positions | |||||
| PIE243 | EST_SSR | 47.67 | No other positions | |||||
| PIE224 | EST_SSR | 42.89 | No other positions | |||||
| WAG020 | EST_SSR | 35.49 | No other positions | |||||
| PIE148 | EST_SSR | 15.73 | No other positions | |||||
| PIE233 | EST_SSR | 12.72 | No other positions | |||||
| FIR023 | EST_SSR | 4.37 | No other positions | |||||
| s_2FT23N_422 | SNP | 70.07 | No other positions | |||||
| s_2FT23N_189 | SNP | 70.07 | No other positions | |||||
| s_AE01EPFEP_360 | SNP | 65.69 | No other positions | |||||
| s_1BABT1_422 | SNP | 65.61 | No other positions | |||||
| s_1AIZHY_768 | SNP | 65.61 | No other positions | |||||
| s_1A5EKO_956 | SNP | 65.61 | No other positions | |||||
| v_9943_223 | SNP | 65.34 | No other positions | |||||
| s_AE01EPFEP_586 | SNP | 65.34 | No other positions | |||||
| s_2IB9FQ_842 | SNP | 65.34 | No other positions | |||||
| s_1APAML_1419 | SNP | 65.34 | No other positions | |||||
| v_9831_1129 | SNP | 65.34 | No other positions | |||||
| s_1AOU7J_489 | SNP | 65.34 | No other positions | |||||
| s_1AHV3Z_964 | SNP | 65.34 | No other positions | |||||
| s_1A0NCP_631 | SNP | 65.33 | No other positions | |||||




