CMap COMPARATIVE MAP VIEWER
The CMap Comparative Map Viewer allows a user to view and compare maps between and among species. See HELP for more information.
 |
Save link
* Bookmarks for this page will fail after this session expires. Use the "Save Link" button to create a permanent link.
General options
** Eliminate Orphans - Remove comparison maps that don't have correspondences to a reference map.
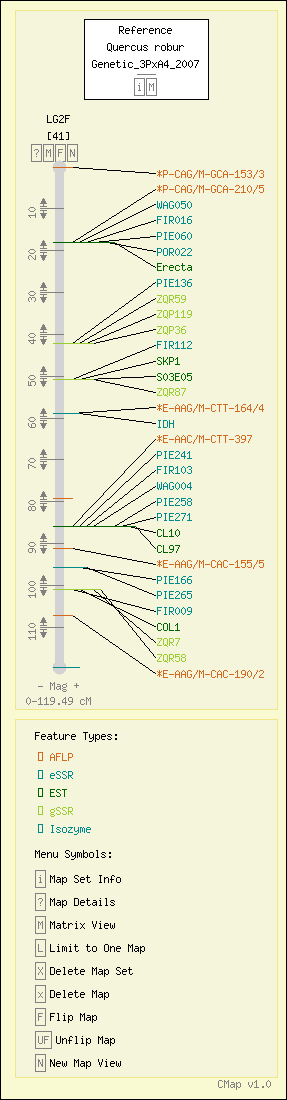
Map Details
| Map Type: | Genetic [ View Map Type Info ] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Map Set Name: | Quercus robur - Genetic_3PxA4_2007 [ View Map Set Info ] | ||||||||||||
| Map Name: | LG2F | ||||||||||||
| Map Start: | 0.00 cM | ||||||||||||
| Map Stop: | 119.49 cM | ||||||||||||
| Features by Type: |
|
Map Features
| [ Download Map Data ]
[ Download Feature Correspondence Data ] |
||||||||
| Quercus robur - Genetic_3PxA4_2007 - LG2F (Click headers to resort) |
Comparative Maps | |||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Type | Position (cM) | Map | Feature | Type | Position | Evidence | Actions |
| ZQR58 | gSSR | 100.95 | Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2M | ZQR58 | gSSR | 84.10 (cM) | Automated name-based | View Maps |
| Quercus robur - Genetic_3PxA4_2009 - LG2M | ZQR58 | gSSR | 27.24 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2009_No_analysis_EST - LG2M | ZQR58 | gSSR | 10.81 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2007 - LG2M | ZQR58 | gSSR | 122.99 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_Robur_Consensus_2011 - LG2 | ZQR58 | gSSR | 34.77 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Leaf_morph3 - LG2M | ZQR58 | gSSR | 84.10 (cM) | Automated name-based | View Maps | |||
| Quercus robur - MultiQTL_3PxA4_2009_Grow_seas - LG2M | ZQR58 | gSSR | 84.10 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Hypoxie - LG2M | ZQR58 | gSSR | 84.10 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2009 - LG2F | ZQR58 | gSSR | 28.17 (cM) | Automated name-based | View Maps | |||
| ZQR7 | gSSR | 100.95 | Quercus robur - MultiQTL_3PxA4_2009_Grow_seas - LG2F | ZQR7 | gSSR | 17.10 (cM) | Automated name-based | View Maps |
| Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2F | ZQR7 | gSSR | 17.10 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Leaf_morph3 - LG2F | ZQR7 | gSSR | 17.10 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus petraea - Genetic_11PxQs29_2011 - LG2M | ZQR7 | gSSR | 75.17 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2009 - LG2M | ZQR7 | gSSR | 24.12 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_Robur_Consensus_2011 - LG2 | ZQR7 | gSSR | 29.84 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2009 - LG2F | ZQR7 | gSSR | 25.69 (cM) | Automated name-based | View Maps | |||
| Quercus petraea - Genetic_Qs28xQs21_2009 - LG2Ma | ZQR7 | gSSR | 7.63 (cM) | Automated name-based | View Maps | |||
| Quercus petraea - Genetic_Qs28xQs21_2011 - LG2F | ZQR7 | gSSR | 49.05 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2007 - LG2M | ZQR7 | gSSR | 122.99 (cM) | Automated name-based | View Maps | |||
| Castanea sativa - QTL_BursaxHoppa_2009_Bud_pheno - LG1F | ZQR7 | gSSR | 43.50 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Leaf_morph1 - LG2F | ZQR7 | gSSR | 17.10 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Leaf_morph2 - LG2F | ZQR7 | gSSR | 17.10 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus petraea - Genetic_11PxQs29_2009 - LG2M | ZQR7 | gSSR | 52.54 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2011 - LG2Fa | ZQR7 | gSSR | 5.15 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus petraea - Genetic_11PxQs29_2009 - LG2F | ZQR7 | gSSR | 21.80 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Hypoxie - LG2F | ZQR7 | gSSR | 17.10 (cM) | Automated name-based | View Maps | |||
| ZQR87 | gSSR | 50.89 | Quercus robur - QTL_3PxA4_2008_Leaf_morph1 - LG2F | ZQR87 | gSSR | 50.70 (cM) | Automated name-based | View Maps |
| Quercus robur - MultiQTL_3PxA4_2009_Grow_seas - LG2M | ZQR87 | gSSR | 43.00 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2F | ZQR87 | gSSR | 24.95 (cM) | Automated name-based | View Maps | |||
| Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2F | ZQR87 | gSSR | 50.80 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Leaf_morph3 - LG2M | ZQR87 | gSSR | 43.00 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Leaf_morph3 - LG2F | ZQR87 | gSSR | 50.80 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2009 - LG2F | ZQR87 | gSSR | 59.63 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Leaf_morph2 - LG2F | ZQR87 | gSSR | 50.70 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Hypoxie - LG2F | ZQR87 | gSSR | 50.80 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2011 - LG2Ma | ZQR87 | gSSR | 61.46 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_Robur_Consensus_2011 - LG2 | ZQR87 | gSSR | 47.71 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2011 - LG2Fb | ZQR87 | gSSR | 44.22 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus petraea - Genetic_11PxQs29_2009 - LG2F | ZQR87 | gSSR | 37.48 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2009_No_analysis_EST - LG2M | ZQR87 | gSSR | 42.20 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2007 - LG2M | ZQR87 | gSSR | 78.95 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2M | ZQR87 | gSSR | 23.21 (cM) | Automated name-based | View Maps | |||
| Quercus robur - MultiQTL_3PxA4_2009_Grow_seas - LG2F | ZQR87 | gSSR | 50.80 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2009 - LG2M | ZQR87 | gSSR | 48.39 (cM) | Automated name-based | View Maps | |||
| Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2M | ZQR87 | gSSR | 43.00 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2M | ZQR87 | gSSR | 23.21 (cM) | Automated name-based | View Maps | |||
| Quercus robur - QTL_3PxA4_2008_Hypoxie - LG2M | ZQR87 | gSSR | 43.00 (cM) | Automated name-based | View Maps | |||
| *P-CAG/M-GCA-153/3 | AFLP | 0.00 | No other positions | |||||
| *E-AAG/M-CAC-190/2 | AFLP | 107.21 | Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2F | *E-AAG/M-CAC-190/2 | AFLP | 11.10 (cM) | Automated name-based | View Maps |
| Quercus robur - MultiQTL_3PxA4_2009_Grow_seas - LG2F | *E-AAG/M-CAC-190/2 | AFLP | 11.10 (cM) | Automated name-based | View Maps | |||
| *P-CAG/M-GCA-87 | AFLP | 119.49 | Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2F | *P-CAG/M-GCA-87 | AFLP | 0.00 (cM) | Automated name-based | View Maps |
| Quercus robur - MultiQTL_3PxA4_2009_Grow_seas - LG2F | *P-CAG/M-GCA-87 | AFLP | 0.00 (cM) | Automated name-based | View Maps | |||
| *P-CAG/M-GCA-210/5 | AFLP | 18.03 | No other positions | |||||
| *E-AAG/M-CTT-164/4 | AFLP | 58.88 | Quercus robur - MultiQTL_3PxA4_2009_Grow_seas - LG2F | *E-AAG/M-CTT-164/4 | AFLP | 43.60 (cM) | Automated name-based | View Maps |
| Quercus robur - Genetic_3PxA4_2009 - LG2F | *E-AAG/M-CTT-164/4 | AFLP | 49.88 (cM) | Automated name-based | View Maps | |||
| Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2F | *E-AAG/M-CTT-164/4 | AFLP | 43.60 (cM) | Automated name-based | View Maps | |||
| *E-AAC/M-CTT-118 | AFLP | 79.33 | Quercus robur - MultiQTL_3PxA4_2009_Debour - LG2F | *E-AAC/M-CTT-118 | AFLP | 26.70 (cM) | Automated name-based | View Maps |
| *E-AAC/M-CTT-397 | AFLP | 86.00 | No other positions | |||||
| *E-AAG/M-CAC-155/5 | AFLP | 91.07 | No other positions | |||||
| *P-CAG/M-GGA-100 | AFLP | 95.61 | No other positions | |||||
| FIR009 | eSSR | 100.95 | No other positions | |||||
| POR023 | eSSR | 119.49 | Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2M | POR023 | eSSR | 58.41 (cM) | Automated name-based | View Maps |
| Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2M | POR023 | eSSR | 58.41 (cM) | Automated name-based | View Maps | |||
| WAG050 | eSSR | 18.03 | No other positions | |||||
| FIR016 | eSSR | 18.03 | No other positions | |||||
| PIE060 | eSSR | 18.03 | No other positions | |||||
| FIR048 | eSSR | 18.03 | No other positions | |||||
| POR022 | eSSR | 18.03 | Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2M | POR022 | eSSR | 3.40 (cM) | Automated name-based | View Maps |
| Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2M | POR022 | eSSR | 3.40 (cM) | Automated name-based | View Maps | |||
| PIE136 | eSSR | 42.27 | Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2M | PIE136 | eSSR | 11.56 (cM) | Automated name-based | View Maps |
| Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2F | PIE136 | eSSR | 12.08 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2F | PIE136 | eSSR | 12.08 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2M | PIE136 | eSSR | 11.56 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2007 - LG2M | PIE136 | eSSR | 60.98 (cM) | Automated name-based | View Maps | |||
| FIR112 | eSSR | 50.89 | No other positions | |||||
| PIE241 | eSSR | 86.00 | Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2F | PIE241 | eSSR | 55.58 (cM) | Automated name-based | View Maps |
| Quercus robur - Genetic_3PxA4_2007 - LG2M | PIE241 | eSSR | 122.99 (cM) | Automated name-based | View Maps | |||
| FIR103 | eSSR | 86.00 | Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2F | FIR103 | eSSR | 58.69 (cM) | Automated name-based | View Maps |
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2F | FIR103 | eSSR | 58.69 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2M | FIR103 | eSSR | 71.52 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2M | FIR103 | eSSR | 71.52 (cM) | Automated name-based | View Maps | |||
| WAG004 | eSSR | 86.00 | Quercus robur - Genetic_3PxA4_2007 - LG2M | WAG004 | eSSR | 122.99 (cM) | Automated name-based | View Maps |
| FIR094 | eSSR | 86.00 | Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2F | FIR094 | eSSR | 68.26 (cM) | Automated name-based | View Maps |
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2F | FIR094 | eSSR | 68.26 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - QTL_EF03xSL03_2009_Leaf_morph - LG2M | FIR094 | eSSR | 61.30 (cM) | Automated name-based | View Maps | |||
| Quercus robur x Quercus robur subsp. slavonica - Genetic_EF03xSL03_2009 - LG2M | FIR094 | eSSR | 61.30 (cM) | Automated name-based | View Maps | |||
| Quercus robur - Genetic_3PxA4_2007 - LG2M | FIR094 | eSSR | 136.42 (cM) | Automated name-based | View Maps | |||




