CMap COMPARATIVE MAP VIEWER
The CMap Comparative Map Viewer allows a user to view and compare maps between and among species. See HELP for more information.
 |
Save link
* Bookmarks for this page will fail after this session expires. Use the "Save Link" button to create a permanent link.
General options
** Eliminate Orphans - Remove comparison maps that don't have correspondences to a reference map.
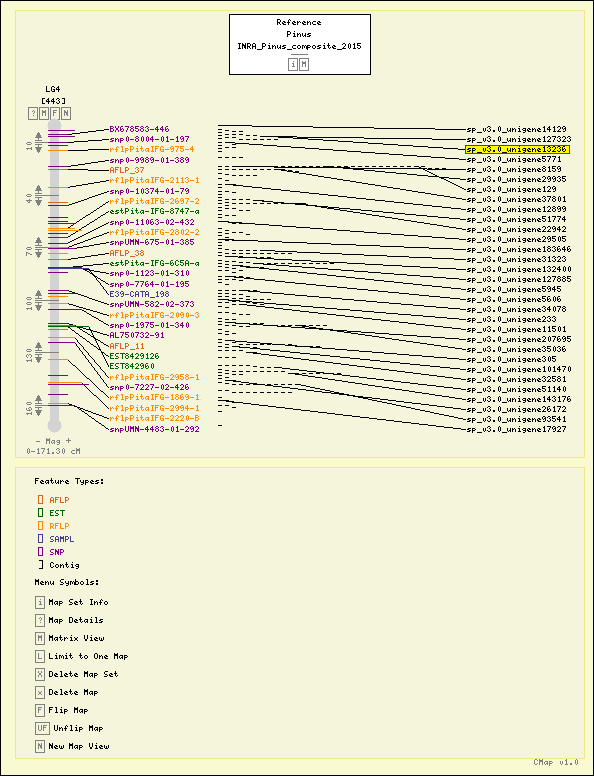
Map Details
| Map Type: | Genetic [ View Map Type Info ] | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Map Set Name: | Pinus - INRA_Pinus_composite_2015 [ View Map Set Info ] | ||||||||||||||
| Map Name: | LG4 | ||||||||||||||
| Map Start: | 0.00 cM | ||||||||||||||
| Map Stop: | 171.30 cM | ||||||||||||||
| Features by Type: |
|
Map Features
| [ Download Map Data ]
[ Download Feature Correspondence Data ] |
||||||||
| Pinus - INRA_Pinus_composite_2015 - LG4 (Click headers to resort) |
Comparative Maps | |||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Type | Position (cM) | Map | Feature | Type | Position | Evidence | Actions |
| sp_v3.0_unigene14129 | Contig | 0.00 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene14129 | Contig | 0.00 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene28227 | Contig | 0.00 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene28227 | Contig | 0.00 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene10469 | Contig | 2.40 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene10469 | Contig | 2.40 (cM) | Automated name-based | View Maps |
| rflpPitaIFG-2145-3 | RFLP | 3.00 | Pinaceae - INRA_Pinaceae_composite_2015 - LG4 | rflpPitaIFG-2145-3 | RFLP | 10.70 (cM) | Automated name-based | View Maps |
| Pinus - INRA_Pinus_composite_2016 - LG4 | rflpPitaIFG-2145-3 | RFLP | 3.00 (cM) | Automated name-based | View Maps | |||
| snp0-3396-01-327 | SNP | 3.00 | Pinus - INRA_Pinus_composite_2016 - LG4 | snp0-3396-01-327 | SNP | 3.00 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene6775 | Contig | 3.00 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene6775 | Contig | 3.00 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene18553 | Contig | 3.60 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene18553 | Contig | 3.60 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene93880 | Contig | 3.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene93880 | Contig | 3.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene26595 | Contig | 4.30 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene26595 | Contig | 4.30 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene1626 | Contig | 5.30 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene1626 | Contig | 5.30 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene2973 | Contig | 5.30 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene2973 | Contig | 5.30 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene44296 | Contig | 5.30 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene44296 | Contig | 5.30 (cM) | Automated name-based | View Maps |
| BX678583-446 | SNP | 5.60 | Pinus - INRA_Pinus_composite_2016 - LG4 | BX678583-446 | SNP | 5.60 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene30092 | Contig | 5.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene30092 | Contig | 5.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene924 | Contig | 5.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene924 | Contig | 5.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene5700 | Contig | 6.10 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene5700 | Contig | 6.10 (cM) | Automated name-based | View Maps |
| snp0-3755-01-387 | SNP | 6.60 | Pinus - INRA_Pinus_composite_2016 - LG4 | snp0-3755-01-387 | SNP | 6.60 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene127323 | Contig | 6.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene127323 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene13236 | Contig | 6.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene13236 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene1906 | Contig | 6.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene1906 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene7046 | Contig | 6.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene7046 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene16720 | Contig | 7.00 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene16720 | Contig | 7.00 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene16767 | Contig | 7.00 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene16767 | Contig | 7.00 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene7769 | Contig | 7.80 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene7769 | Contig | 7.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene129391 | Contig | 8.10 | Pinus - INRA_Pinus_composite_2016 - LG4 | sp_v3.0_unigene129391 | Contig | 8.10 (cM) | Automated name-based | View Maps |




