CMap COMPARATIVE MAP VIEWER
The CMap Comparative Map Viewer allows a user to view and compare maps between and among species. See HELP for more information.
Use  and
and  below to scroll image horizontally
below to scroll image horizontally
 and
and  below to scroll image horizontally
below to scroll image horizontally|
|
|
|
|
Save link
* Bookmarks for this page will fail after this session expires. Use the "Save Link" button to create a permanent link.
General options
** Eliminate Orphans - Remove comparison maps that don't have correspondences to a reference map.
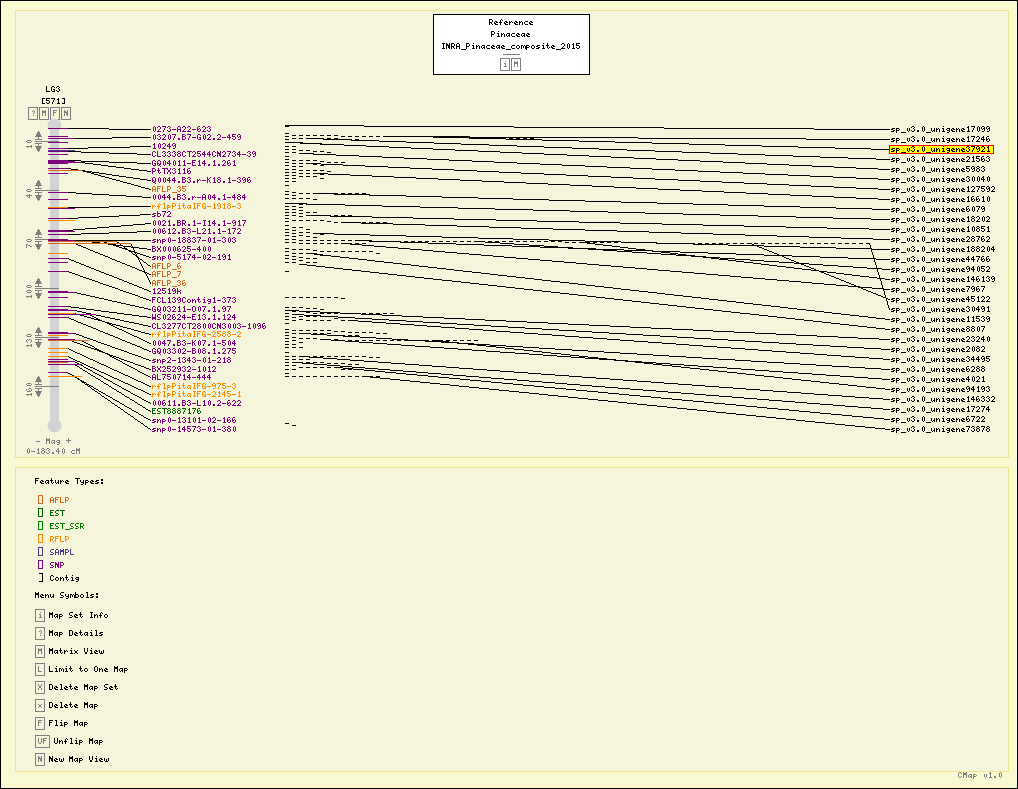
Map Details
| Map Type: | Genetic [ View Map Type Info ] | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Map Set Name: | Pinaceae - INRA_Pinaceae_composite_2015 [ View Map Set Info ] | ||||||||||||||||
| Map Name: | LG3 | ||||||||||||||||
| Map Start: | 0.00 cM | ||||||||||||||||
| Map Stop: | 183.40 cM | ||||||||||||||||
| Features by Type: |
|
Map Features
| [ Download Map Data ]
[ Download Feature Correspondence Data ] |
||||||||
| Pinaceae - INRA_Pinaceae_composite_2015 - LG3 (Click headers to resort) |
Comparative Maps | |||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Type | Position (cM) | Map | Feature | Type | Position | Evidence | Actions |
| AFLP_8 | AFLP | 132.00 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | AFLP_8 | AFLP | 132.00 (cM) | Automated name-based | View Maps |
| A6D12 | EST_SSR | 71.10 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | A6D12 | EST_SSR | 71.10 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene17099 | Contig | 0.00 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene17099 | Contig | 0.00 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene29834 | Contig | 1.00 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene29834 | Contig | 1.00 (cM) | Automated name-based | View Maps |
| 0273-A22-623 | SNP | 2.30 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | 0273-A22-623 | SNP | 2.30 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene7534 | Contig | 5.30 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene7534 | Contig | 5.30 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene17255 | Contig | 6.20 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene17255 | Contig | 6.20 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene17990 | Contig | 6.20 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene17990 | Contig | 6.20 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene18583 | Contig | 6.20 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene18583 | Contig | 6.20 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene210367 | Contig | 6.20 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene210367 | Contig | 6.20 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene28548 | Contig | 6.20 | Pinaceae - INRA_Pinaceae_composite_2015 - LG2 | sp_v3.0_unigene28548 | Contig | 59.90 (cM) | Automated name-based | View Maps |
| Pinaceae - INRA_Pinaceae_composite_2016 - LG2 | sp_v3.0_unigene28548 | Contig | 59.90 (cM) | Automated name-based | View Maps | |||
| Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene28548 | Contig | 6.20 (cM) | Automated name-based | View Maps | |||
| sp_v3.0_unigene23523 | Contig | 6.60 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene23523 | Contig | 6.60 (cM) | Automated name-based | View Maps |
| BX000625-400 | SNP | 71.10 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | BX000625-400 | SNP | 71.10 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene7240 | Contig | 6.70 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene7240 | Contig | 6.70 (cM) | Automated name-based | View Maps |
| 5720 | SNP | 6.80 | No other positions | |||||
| sp_v3.0_unigene25054 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene25054 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene29549 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene29549 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene12133 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene12133 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene137679 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene137679 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene145182 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene145182 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene147036 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene147036 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene16539 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene16539 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene17246 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene17246 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene17440 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene17440 | Contig | 6.80 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene20237 | Contig | 6.80 | Pinaceae - INRA_Pinaceae_composite_2016 - LG3 | sp_v3.0_unigene20237 | Contig | 6.80 (cM) | Automated name-based | View Maps |