CMap COMPARATIVE MAP VIEWER
The CMap Comparative Map Viewer allows a user to view and compare maps between and among species. See HELP for more information.
 |
Save link
* Bookmarks for this page will fail after this session expires. Use the "Save Link" button to create a permanent link.
General options
** Eliminate Orphans - Remove comparison maps that don't have correspondences to a reference map.
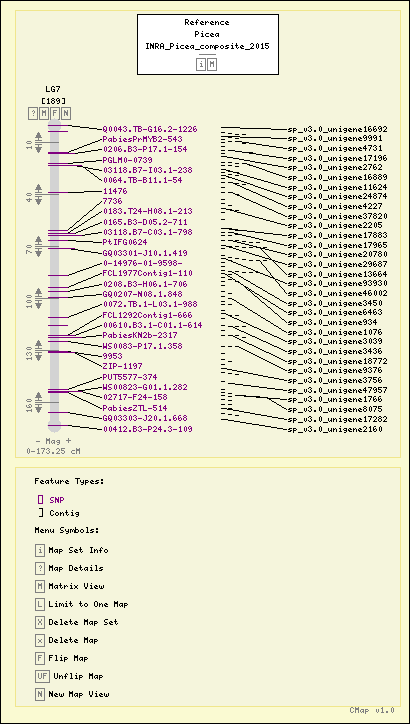
Map Details
| Map Type: | Genetic [ View Map Type Info ] | ||||||
|---|---|---|---|---|---|---|---|
| Map Set Name: | Picea - INRA_Picea_composite_2015 [ View Map Set Info ] | ||||||
| Map Name: | LG7 | ||||||
| Map Start: | 0.00 cM | ||||||
| Map Stop: | 173.25 cM | ||||||
| Features by Type: |
|
Map Features
| [ Download Map Data ]
[ Download Feature Correspondence Data ] |
||||||||
| Picea - INRA_Picea_composite_2015 - LG7 (Click headers to resort) |
Comparative Maps | |||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Type | Position (cM) | Map | Feature | Type | Position | Evidence | Actions |
| sp_v3.0_unigene5812 | Contig | 27.43 | No other positions | |||||
| sp_v3.0_unigene28012 | Contig | 27.57 | No other positions | |||||
| sp_v3.0_unigene26751 | Contig | 28.80 | No other positions | |||||
| sp_v3.0_unigene24874 | Contig | 29.22 | No other positions | |||||
| sp_v3.0_unigene432 | Contig | 29.37 | No other positions | |||||
| sp_v3.0_unigene6930 | Contig | 30.28 | No other positions | |||||
| sp_v3.0_unigene5757 | Contig | 30.95 | No other positions | |||||
| sp_v3.0_unigene23555 | Contig | 33.16 | No other positions | |||||
| sp_v3.0_unigene4227 | Contig | 33.17 | No other positions | |||||
| sp_v3.0_unigene12584 | Contig | 33.64 | Picea - INRA_Picea_composite_2015 - LG9 | sp_v3.0_unigene12584 | Contig | 123.02 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene7503 | Contig | 34.39 | No other positions | |||||
| sp_v3.0_unigene4228 | Contig | 35.81 | No other positions | |||||
| sp_v3.0_unigene15164 | Contig | 38.93 | No other positions | |||||
| sp_v3.0_unigene37820 | Contig | 39.08 | No other positions | |||||
| 11476 | SNP | 39.18 | No other positions | |||||
| sp_v3.0_unigene29937 | Contig | 40.11 | No other positions | |||||
| sp_v3.0_unigene7966 | Contig | 43.27 | No other positions | |||||
| sp_v3.0_unigene6942 | Contig | 45.45 | No other positions | |||||
| sp_v3.0_unigene94157 | Contig | 46.32 | No other positions | |||||
| sp_v3.0_unigene2205 | Contig | 47.96 | No other positions | |||||
| sp_v3.0_unigene17644 | Contig | 48.74 | No other positions | |||||
| sp_v3.0_unigene24360 | Contig | 49.29 | Picea - INRA_Picea_composite_2015 - LG10 | sp_v3.0_unigene24360 | Contig | 108.14 (cM) | Automated name-based | View Maps |
| sp_v3.0_unigene34067 | Contig | 50.28 | No other positions | |||||
| sp_v3.0_unigene8869 | Contig | 54.40 | No other positions | |||||
| sp_v3.0_unigene17883 | Contig | 59.61 | No other positions | |||||




